Oct 31 2018
A team of scientists from the University of Illinois at Urbana-Champaign and Mayo Clinic have designed a unique type of molecular probe that can measure and count RNA in cells and tissue without organic dyes. The probe is based on the conventional fluorescence in situ hybridization (FISH) method, but it depends on compact quantum dots to illuminate molecules and diseased cells instead of fluorescent dyes.
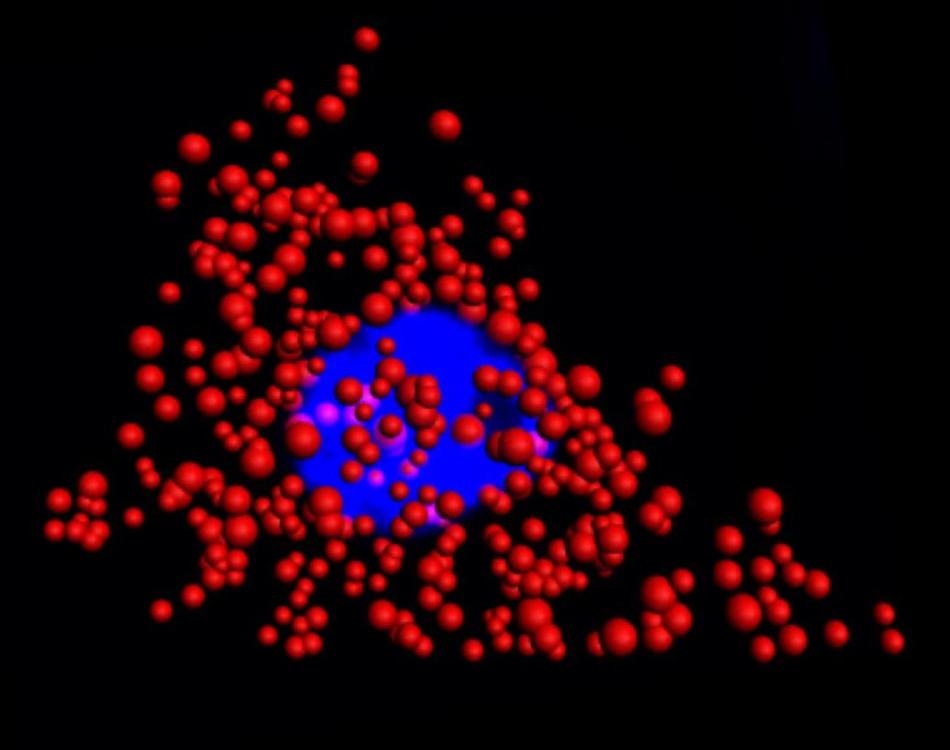 Quantum dots illuminate the locations of individual mRNA as red dots in the cytoplasm of a single HeLa cell. The blue region is the nucleus. This work was a collaborative effort between Illinois Bioengineering and Mayo Clinic researchers. (Credit: University of Illinois at Urbana-Champaign)
Quantum dots illuminate the locations of individual mRNA as red dots in the cytoplasm of a single HeLa cell. The blue region is the nucleus. This work was a collaborative effort between Illinois Bioengineering and Mayo Clinic researchers. (Credit: University of Illinois at Urbana-Champaign)
Over the last five decades, FISH has advanced into a multi-billion-dollar industry because it successfully images and counts RNA and DNA in single cells. However, FISH has its limits because of the delicate nature of the dyes. For instance, the dyes quickly deteriorate and are not very efficient at imaging in three dimensions. Furthermore, conventional FISH can only read out a couple of DNA or RNA sequences at a time.
By replacing dyes with quantum dots, there are no stability issues whatsoever and we can count numerous RNAs with higher fidelity than before. Moreover, we uncovered a fundamental limit to the size of a molecular label in cells, revealing new design rules for analysis in cells.
Andrew Smith, associate professor of Bioengineering and member of the research team
In their recent paper, published October 26th, 2018, in the online edition of Nature Communications, Smith and his team identified an ideal size for quantum dots that would effectively work with the FISH protocol. This finding enabled quantum dot-based FISH to match the labeling accuracies presently obtained with organic dyes.
The team developed unique quantum dots that are composed of a zinc, cadmium, selenium, and mercury alloy and are coated with polymers. “The core of the dot dictates the wavelength of emission, and the shell dictates how much light will be given off,” said Smith, who is also affiliated with the Micro + Nanotechnology Lab, Carle Illinois College of Medicine, and Department of Materials Science and Engineering at the University of Illinois.
Unlike the conventional quantum dots, these dots can release color autonomous of the size of the particle. The dots are also sufficiently small (7 nm) to fit on a probe that can move between proteins and DNA in a cell, rendering them more comparable in size to the dyes used in conventional FISH probes.
In experiments with HeLa cells and prostate cancer cells, the scientists discovered that dye-based FISH cell counts diminished quickly in minutes. The quantum dot-based FISH technique provided long-term luminescence to permit counting of RNA for over 10 minutes, making it possible to obtain 3D cell imaging.
This is important because images of cells and tissues are acquired slice-by-slice in sequence, so later slices that are labeled with dyes are depleted before they can be imaged.
Andrew Smith
This study is part of the Mayo Illinois Alliance in which engineers from Illinois work directly with clinicians and biologists from Mayo Clinic to address unresolved medical challenges. The Mayo Clinic Biomarker Discovery Group is aiming to create FISH-based diagnostics for tumor biopsies so as to enhance the accuracy of cancer diagnosis, to select tailored treatments, and to enhance prognoses. The QD-FISH methodology was formulated to meet this need for biomarker panels necessitating the analysis of many genetic changes in cells in tumors for which only small amounts of specimens may be available when obtained with a fine needle, as is typically the case in prostate cancer.
This work was supported by the Mayo-Illinois Alliance, National Institutes of Health, and the National Science Foundation. The full title of the paper is "Enhanced mRNA FISH with Compact Quantum Dots."
Besides Smith, the following Illinois Bioengineering researchers contributed to the research: Yang Liu, Phuong Le, Sung Jun Lim, Liang Ma, and Suresh Sarkar. Mayo Clinic collaborators include: Dr. Farhad Kosari, Stephen Murphy, John Cheville, and George Vasmatzis.